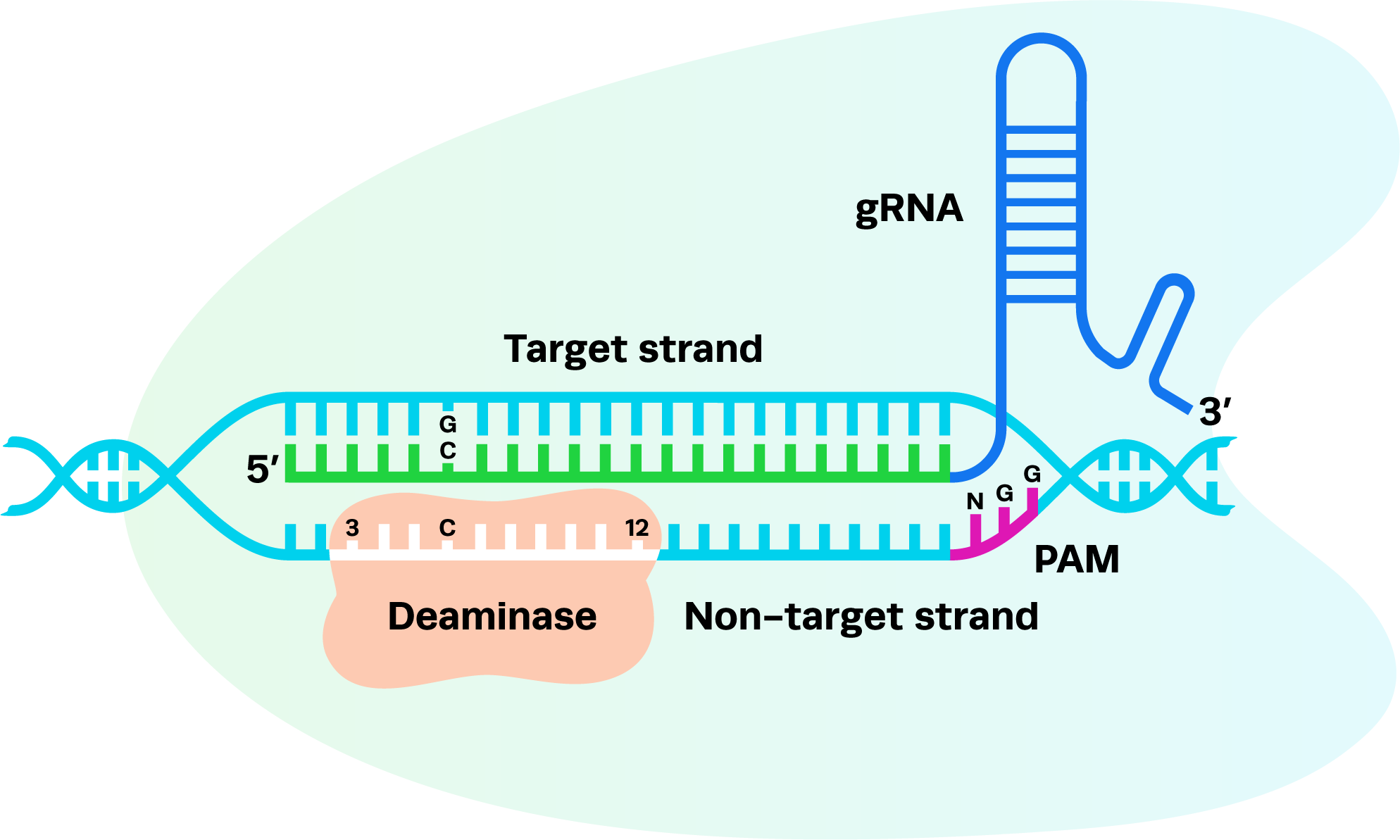
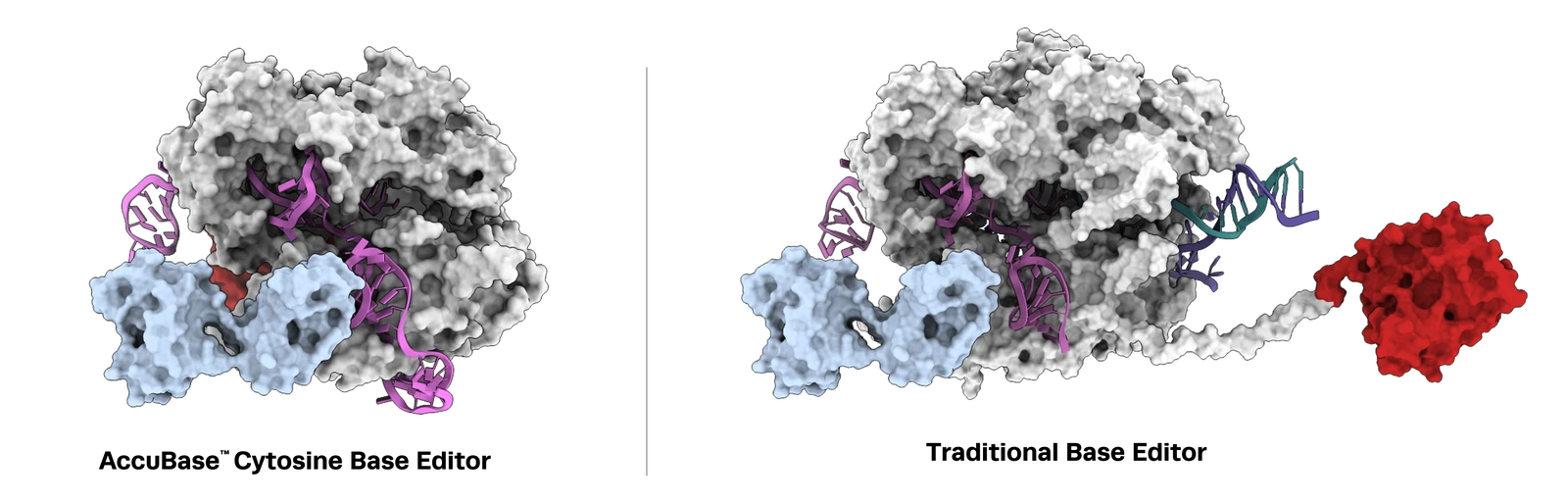
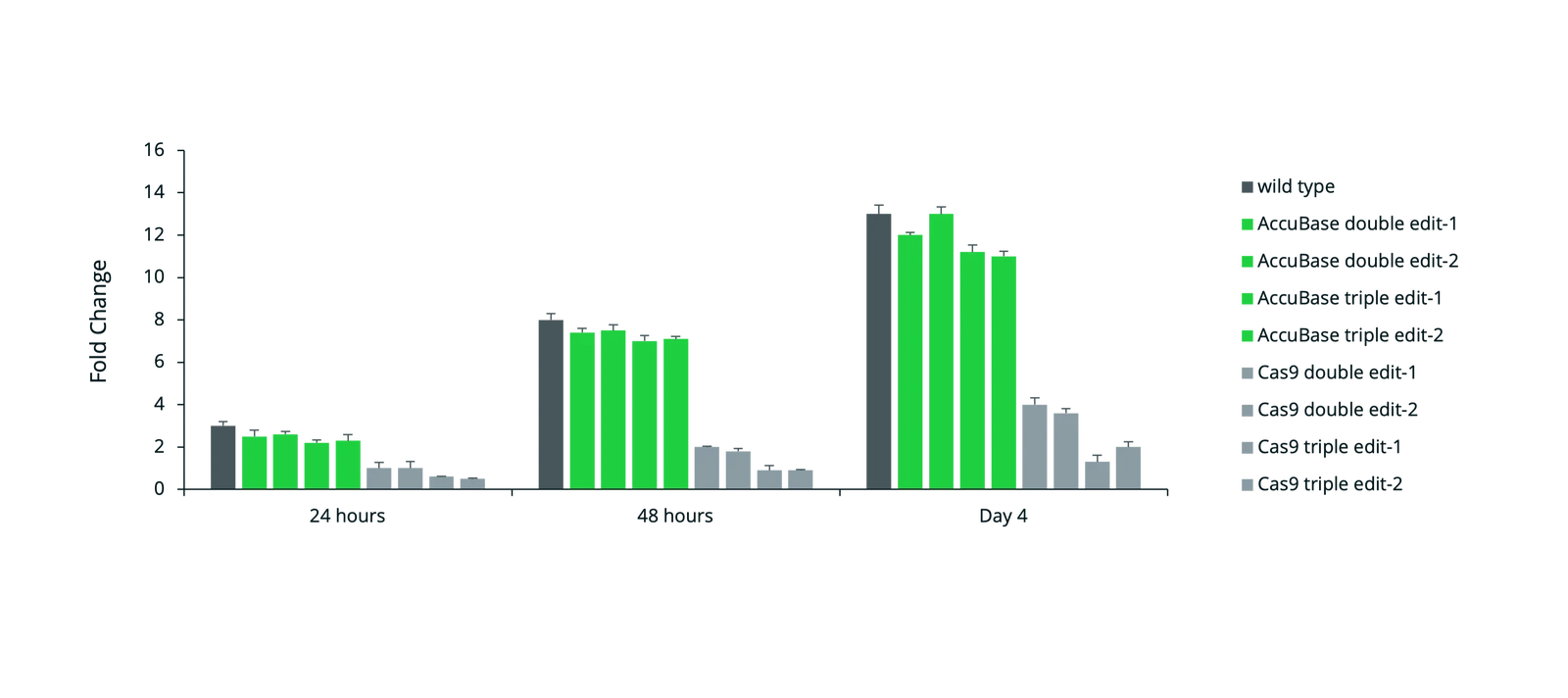
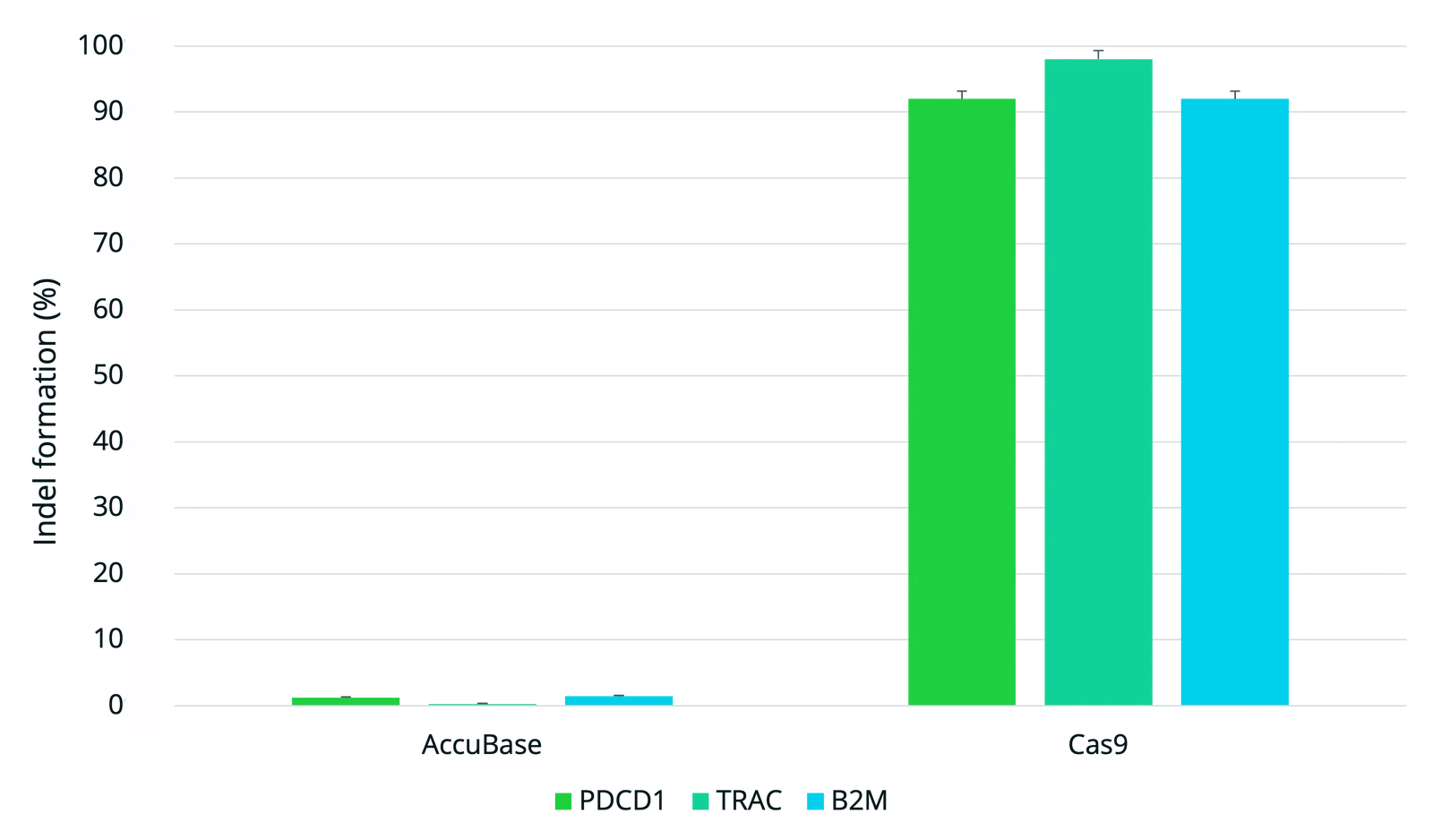
For AccuBase systems, the editing window spans nucleotides 3 to 12 (with position 1 being furthest from the 5'-NGG-3' PAM). The target cytosine must ideally fall within this editing window to maximize active editing while reducing unintended modifications. When selecting binding sites:
- Ensure PAM proximity: Choose a 5’-NGG-3’ PAM sequence that aligns the target cytosine centrally within the editing window.
- Avoid boundary targets: Cytosine bases outside this editing range may have significantly reduced editing efficiency.
- Evaluate bystander cytosines: If multiple cytosines fall within or near the editing window, prioritize designs that limit edits to irrelevant or non-critical regions to avoid unintentional consequences.
To demonstrate the editing window of AccuBase, the image below highlights positions where C-to-T base conversions could occur using our AccuBase positive control gRNA as examples.