-
gRNA components. Most gRNA molecules have two components: the target sequence that hybridizes with the genomic target and a constant sequence, or scaffold sequence, that enables the gRNA to complex with the nuclease. When we discuss gRNA design here, we will focus on the target sequence.
-
PAM availability. Most nucleases have a unique protospacer adjacent motif (PAM) sequence. For example, SpCas9 has an NGG PAM. The availability of gRNA target sequences in your region of interest will depend on the presence and location of PAM sequences in your target region. This chapter of our How to Use CRISPR guide explains the importance of the PAM sequence.
-
Specificity. A good gRNA will guide your nuclease to create high-efficiency edits at the intended target site (on-target) while minimizing editing at other sites (off-target). Most design tools provide information about the predicted off-target sites for each suggested gRNA design. This blog discusses the significance of off-target editing in CRISPR experiments and methods for predicting or detecting these undesirable edits.
-
Target site location. The ideal target site will depend on your experimental application. Before starting the gRNA design process, consider your experimental needs.
-
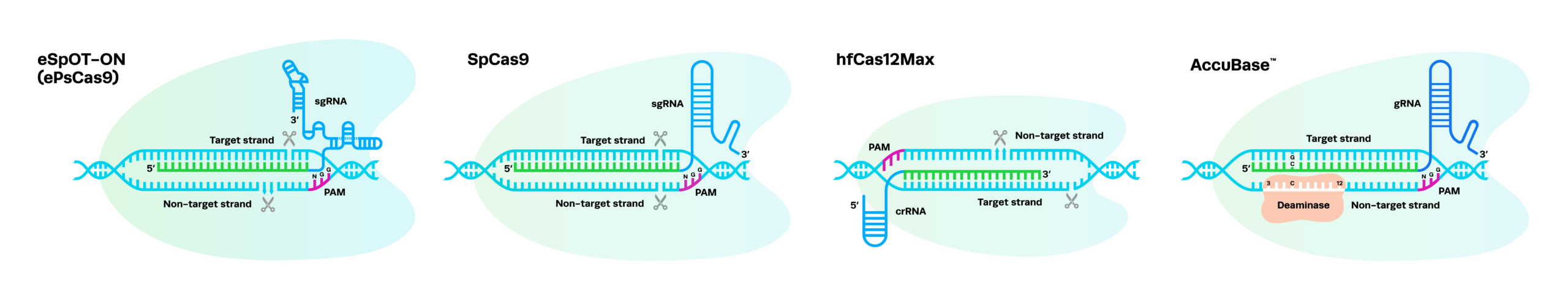
CRISPR nucleases (eSpOT-ON, hfCas12Max, and SpCas9) that form double-strand breaks (DSBs) are frequently used for knockout and knock-in edits.
-
Knockout edit: We recommend creating a cut site in an early exon shared by as many transcripts as possible. This will maximize your chances of creating an early frameshift to disrupt gene expression.
-
Knock-in edit: We recommend selecting gRNAs with target sites as close to the site of your intended insertion as possible (creating a cut site <10 bp away from the intended insertion is ideal).
-
-
Base editors (AccuBase) create base conversions within a specific window relative to the gRNA target site.
-
AccuBase creates cytosine (C) to thymine (T) base edits within a 3-12 base editing window, where position 1 is furthest from the PAM. Within the window, the rate of C to T conversions is highest in positions 7-11.
-
-
For most applications, we recommend targeting exons shared by as many transcripts as possible. This will ensure a functional edit across isoforms. Databases like Ensembl can help you identify suitable exons for this purpose.
-
-
Other considerations. When selecting your gRNAs, you may wish to account for parameters that are not discussed here. These include the predicted activity of your gRNA and other experiment-specific considerations. For CRISPR experiments beyond knockout, knock-in, and base edits, we recommend reviewing our CRISPR gRNA design for diverse applications resource or contacting our technical support team for guidance.
Learn more
Webcast featuring eSpOT-ON
Explore More
eSpOT-ON Nuclease Protein Available Now
Explore More
Order eSpOT-ON Nuclease mRNA Now