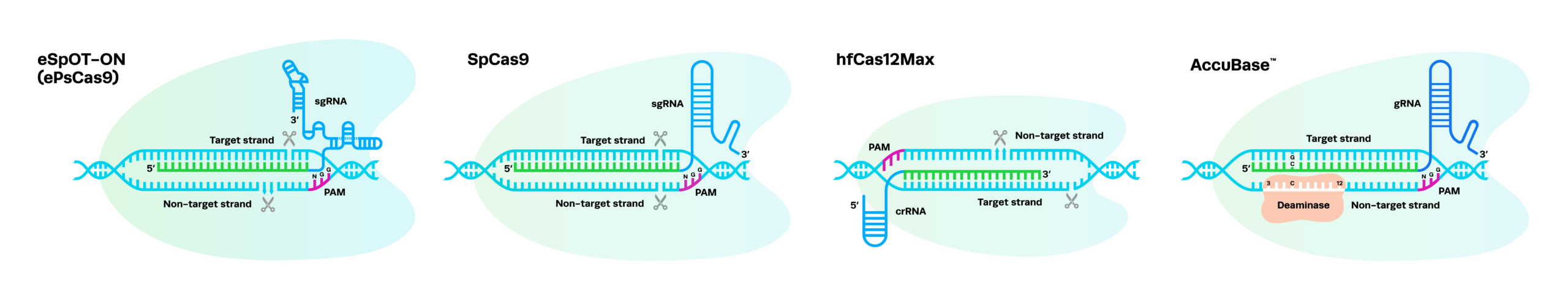
For any CRISPR genome editing experiment, you need the two core components of the CRISPR editing machinery: CRISPR enzyme or Cas nuclease (CRISPR-associated endonuclease) and guide RNA (gRNA).
Learn more
Webcast featuring eSpOT-ON
Explore More
eSpOT-ON Nuclease Protein Available Now
Explore More
Order eSpOT-ON Nuclease mRNA Now